-Search query
-Search result
Showing 1 - 50 of 98 items for (author: rosenthal & pb)

EMDB-17001: 
In-situ structure of the hexameric HEF trimers from influenza C viral particles
Method: subtomogram averaging / : Liu ZB, Rosenthal PB
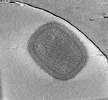
EMDB-18916: 
Cryotomogram of mature Vaccinia virus (WR) virion
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18917: 
Subtomogram average of the Vaccinia virus (WR) portal complex in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-18918: 
Subtomogram average of the Vaccinia virus (WR) A4/A10 palisade trimer in mature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-16150: 
8:1 binding of FcMR on IgM pentameric core
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-16151: 
FcMR binding at subunit Fcu1 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-16152: 
FcMR binding at subunit Fcu3 of IgM pentamer
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15596: 
In situ subtomogram average of Vaccinia virus (WR) palisade, all virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15597: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from CEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15598: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IMVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15599: 
In situ subtomogram average of Vaccinia virus (WR) palisade, from IEVs
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15600: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (immature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15601: 
Cryotomogram of Vaccinia virus (WR) infected HeLa cell (mature virions)
Method: electron tomography / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15602: 
In situ subtomogram average of Vaccinia virus (WR) D13 lattice, on immature virions
Method: subtomogram averaging / : Calcraft T, Hernandez-Gonzalez M, Nans A, Rosenthal PB, Way M

EMDB-15181: 
Prefusion spike of SARS-CoV-2 (Wuhan), closed conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB
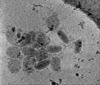
EMDB-15182: 
Electron cryotomography of SARS-CoV-2 virions
Method: electron tomography / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15183: 
Whole SARS-CoV-2 (Wuhan) virion
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-15185: 
Prefusion spike of SARS-CoV-2 (Wuhan), 1-RBD-up conformation
Method: subtomogram averaging / : Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB

EMDB-13921: 
Cryo-EM structure of full-length human immunoglobulin M
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-13922: 
Cryo-EM structure of human monomeric IgM-Fc
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15375: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15376: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 2
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15377: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 3
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15379: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-15380: 
Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 4
Method: single particle / : Chen Q, Rosenthal P, Tolar P

EMDB-14742: 
X-31 Hemagglutinin Precursor HA0 at pH 7.5
Method: single particle / : Garcia-Moro E, Rosenthal PB

EMDB-14743: 
X-31 Hemagglutinin Precursor HA0 at pH 4.8
Method: single particle / : Garcia-Moro E, Rosenthal PB

EMDB-14744: 
X-31 Hemagglutinin Precursor HA0 at pH 7.5 after reneutralization
Method: single particle / : Garcia-Moro E, Rosenthal PB

EMDB-14225: 
Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement)
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14227: 
Dissociated S1 domain of Beta Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement)
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14228: 
Dissociated S1 domain of Mink Variant SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement)
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14229: 
Alpha Variant SARS-CoV-2 Spike in Closed conformation
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14230: 
Alpha Variant SARS-CoV-2 Spike with 1 Erect RBD
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14232: 
Beta Variant SARS-CoV-2 Spike with 1 Erect RBD
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14233: 
Beta Variant SARS-CoV-2 Spike with 2 Erect RBDs
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14234: 
Mink Variant SARS-CoV-2 Spike in Closed conformation
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14235: 
Mink Variant SARS-CoV-2 Spike with 2 Erect RBDs
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14237: 
Mink Variant SARS-CoV-2 Spike with 1 Erect RBD
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14226: 
Dissociated S1 domain of Alpha Variant SARS-CoV-2 Spike bound to ACE2
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14231: 
Alpha Variant SARS-CoV-2 Spike with 2 Erect RBDs
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-14236: 
Furin Cleaved Alpha Variant SARS-CoV-2 Spike in complex with 3 ACE2
Method: single particle / : Benton DJ, Wrobel AG, Gamblin SJ

EMDB-12585: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12586: 
Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect)
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-12587: 
Trimeric SARS-CoV-2 spike ectodomain bound to P008_056 Fab
Method: single particle / : Rosa A, Pye VE, Nans A, Cherepanov P

EMDB-11928: 
SctV (SsaV) cytoplasmic domain
Method: single particle / : Matthews-Palmer TRS, Gonzalez-Rodriguez N, Calcraft T, Lagercrantz S, Zachs T, Yu XJ, Grabe G, Holden D, Nans A, Rosenthal P, Rouse S, Beeby M

EMDB-10810: 
In-situ structure of the trimeric HEF from influenza C viral particles with a matrix layer by cryo-ET
Method: subtomogram averaging / : Halldorsson S, Rosenthal PB

EMDB-10811: 
In-situ structure of a dimer of the trimeric HEF from influenza C viral particles containing a matrix layer by cryo-ET
Method: subtomogram averaging / : Halldorsson S, Rosenthal PB

EMDB-10812: 
In-situ structure of the trimeric HEF from influenza C viral particles without a matrix layer by cryo-ET
Method: subtomogram averaging / : Halldorsson S, Rosenthal PB

EMDB-10813: 
In-situ structure of a dimer of the trimeric HEF from influenza C viral particles without a matrix layer by cryo-ET
Method: subtomogram averaging / : Halldorsson S, Rosenthal PB

EMDB-10814: 
In-situ structure of HEF from filamentous influenza C viral particles by cryo-ET
Method: subtomogram averaging / : Halldorsson S, Rosenthal PB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
